By DeeAnn Visk and Nisha Nair
Maimonides, a twelfth century physician, wrote a daily prayer:
“Let the thought never arise that I have
Attained enough knowledge
But vouchsafe to me ever the strength,
The leisure and eagerness to add to what I know
For art is great and the mind of man ever growing.”
This clearly sums up the attitude of physicians toward continuing medical education in a bygone era. With the continuing advances in the field of medicine and diagnostics, practitioners and other health professionals need to remain current on updates regarding recent advances in their field. Most physicians and medical researchers have not had formal training in CME writing. Therefore, there is an ever-increasing need for medical writers to effectively disseminate knowledge by clearly communicating progress in medicine. Hence, the NorCal AMWA chapter addressed the need for CME writers during a recent meeting.

Introduction to CME as a Medical Writer
The talk was presented by Joshua Schechtel, MD, MPH, FAAP, guest speaker. Josh is a self-employed consultant with 25 years of experience in the field of CME writing and is currently the Manager of the Medical Education Program for San Mateo County Health. Previously he was the medical director of the Permanente medical group, a member of the editorial advisory board of the New Physician magazine, and has served on the board of the Pacific Horticultural Society for 12 years.
Josh spoke at length about the theory and practice of CME as it exists today. He also provided an overview of the history of CME in the US and how various regulations have evolved to make the CME industry what it is today. In order to understand the opportunities for medical writers in continuing medical education (CME), it is first necessary to understand some basics about CME.
Water lilies. Photo by Joshua Schechtel
Conflicts of Interest, Real and Perceived
Josh also outlined the regulations that govern disclosure of conflicts of interest, both technical and financial, of the author, an important step for anyone who makes a foray into the field of CME writing.
One of the most important elements in understanding CME is the importance of disclosures. Josh began his presentation by noting that he does “not have a financial interest/arrangement or affiliation with any organization that could be perceived as a real or apparent conflict of interest in the context of the subject of this presentation.”
Any reputable offering of CME must disclose any conflict of interest as well as anything that could be construed as a perceived conflict of interest. These conflicts of interest range from stock in a company to involvement in clinical trials of a new drug. Even if the conflict could merely be construed, it must be disclosed.
An example of a perceived conflict can be found in a situation where a practicing psychiatrist was presenting a CME about screening for depression. The psychiatrist had received grants for drug research from pharmaceutical companies that market drugs for depression. This may not be considered a potential conflict of interest, as the research grant specifically funded treatment for depression and did not fund research for tools used in diagnosis of depression. Hence, it was fine for this psychiatrist to present a CME on screening for depression, which is considered separate and distinct from treating depression.
Most conflicts of interest do not preclude medical doctors from presenting CME, as long as they are disclosed upfront. This is analogous to the situation in publishing papers. Authors with conflicts of interest can publish findings; they simply need to disclose real or perceived conflicts of interest.
Embarcadero, San Francisco, CA, near to where the NorCalAMWA meeting was held
Medical Writers Generally Have No Conflicts of Interests
During the question and answer section, the point of medical writers having conflicts of interest arose. In general, most medical writers will not have a conflict of interest if they have written for a CME organization and want to take work at a different CME entity.
Each State Has a Body to Oversee CME for that Particular State
Another key point was the understanding that the individual states have State Medical Boards that govern the practice of medicine in that state; the State of California was the example used for this meeting. If writing CME for another state, then refer to the rules for the medical board of that particular state. This is mainly to ensure unbiased information. In some states, the rules are very explicit but in California, the regulations are more implied to ensure transparency.
The California Legislature defines CME as educational activities that meet the standards for the Division of Licensing and serve to maintain, develop or increase the knowledge, skills, and professional performance that physicians or surgeons use to provide care, or improve the quality of care provided for patients, including, but not limited to, educational activities that meet any of the following criteria:
- “Have a scientific or clinical content with a direct bearing on the quality or cost-effective provision of patient care, community or public health, or preventive medicine
- Concern quality assurance or improvement, risk management, health facility standards, or the legal aspects of clinical medicine
- Concern bioethics or professional ethics
- Designed to improve the physician/patient relationship”
The definition expressly excludes: “educational activities that are not directed toward the practice of medicine, or are directed toward the business aspects of medical practice, including, but not limited to, medical office management, billing and coding, and marketing.”
Details of rotunda in the California State Legislature building
What CME is Not
To prevent frivolous offerings of CME, the following are NOT allowed for CME, as they are neither education activities, nor directed towards the practice of medicine:
- “Medical office management in integrated healthcare delivery/group practice arrangements
- Marketing of integrated delivery systems/group practice arrangements
- Understanding corporate structure from a financial or legal perspective”
All CME providers must be accredited to provide CME in a particular state.
Who Accredits Organizations that Offer CME?
Historically, the American Medical Association (AMA) published the Physicians Recognition Award and Credit system and trademarked the term AMA PRA Category 1 CreditTM. Legislative reform attempt to update and make it current for those practicing medicine
Accredited CME providers may certify nonclinical subjects (such as office management, patient-physician communication). The line gets rather grey here; several of these subjects may be rather questionable.
The institute for Medical Quality (IMQ)/California Medical Association (CMA) is recognized by the Accreditation Council of Continuing Medical Education. This means that all organizations wising to offer CME in the State of California, must first be accredited by IMQ/CMA. Thus, medical writers developing a relationship with CME offering organizations, should check to see if the organization is accredited by IMQ/CMA.
A Brief History of CME in the US
The earliest CME was probably in the 1920’s at the Mayo Clinic. In 1927 the Bulletin of the Mayo Clinic and the Mayo Foundation began publishing; this publication evolved into the present Mayo Clinic Proceedings. Before this time, physicians found it difficult to keep current with new practices.
The first medical specialty organization in the US, the American Urological Association, had the first mandatory CME program in 1934. In 1957, the American Medical Association (AMA) published the first CME guidelines. By 1968 that evolved into the physician recognition award. The Accreditation Council for Continuing Medical Education was created in 1981 as a way to lessen AMA’s hold on CME.
In 2007, the Senate Committee on Finance questioned the pharmaceutical industry’s funding of CME for physicians. This report led to reforms in the system, but constant vigilance is still needed.
Careers in CME Writing
Much CME content is written by medical providers; unfortunately, most clinicians are untrained in the art of medical writing, Nonetheless, they are content experts. Medical writers contribute to the generation of excellent CME by writing content and editing drafts. Clinicians and medical writers work together to address the needs of physicians to stay abreast of current medical best practices. Practicing clinicians are subject matter experts, providing expertise on CME content. Medical writers ensure the accuracy and readability of CME content from a writing standpoint, with appropriate citations as needed.
Photo taken while traveling from San Francisco to San Diego. Photo by DeeAnn Visk.
Trends in CME writing
Josh analyzed trends over the past 10 to 12 years, using the ACCME 2017 Data Report.
- Accredited providers reported close to $2.7 B in investment in medical education from a variety of sources in 2017, an increase of 6% from 2016.
- 44,000 activities for 105,000 hours of interaction, with 6 M physician interaction and millions of other learner interactions; non-physician accounted for about 7.8 M interactions
- There are about 1800 certified providers offering accredited CME in the US
- About 163,000 courses are offered for CME nation-wide.
- 2005 to 2017 witnessed an internet-based CME increased about 7-fold.
- For non-physicians there was an 8-fold increase from 2006 to 2016.
- A much larger audience for internet-based materials than print journals was noted.
A quick Google search for “continuing medical education online” returned 500 million results, reinforcing the large number of offerings that are web-based.
Here are a few of the most familiar organizations providing CME:
To find jobs/work/projects for medical writing, places to look include:
Types of CME Writing Assignments
- Slide decks
- Monographs or journal supplements
- Online clinical case studies
- Video round table discussion, more conversational style
- Audio or video podcast
- Newsletters
Visiting Lassen Volcanic National Park does not count as CME. Photo by DeeAnn Visk.
Questions and Answers
The discussion following the presentations covered a wide range of topics.
Pharmaceutical companies discover many new and effective drugs. In order to train specialists to utilize the drugs effectively, the companies may offer CME programs in order to train physicians on how to best to use them. New approaches may also need a fresh diagnostic approach. Hence, what can appear to be a blatant conflict of interest, should be considered in the light of educating doctors, who may be unaware of new developments. Balancing this need for further education with the for-profit motive can be challenging. Medical writers need to carefully consider this before determining if a conflict of interest exists and while preparing a particular CME proposal.
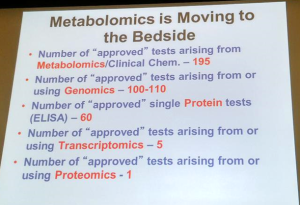
Additionally, new therapeutic approaches often require the use of diagnostic tests to correctly identify patients for whom the treatment is effective. Conversely, a diagnostic test may also be used to rule out a specific therapy for patient treatment. Diagnostic/prognostic tests can include patient questionnaires, blood analytes, genetic sequencing (both somatic and germline), imaging, and staining of tissue sections. Training doctors to correctly order and interpret the tests for new therapies requires instruction—often in the form of CME.
Another topic of interest was the “needs assessment.” Basically, this is a rationale for why a particular CME needs to be offered. The needs assessment usually includes a polling of clinicians, to determine the necessity of preparing CME materials for a specific topic. The national AMWA website offers more insights into writing needs assessments.
Conclusion
In short, writing CME is more of a RIGHT-ing job, defining the need and bridging the gap in an existing specialty. CME writing is all about contributing to healthcare in the WRITE way; it requires conscientious research, precise writing, and devotion to accuracy. Medical writers are an important part of the team, working towards the goal of informing clinicians on updates to emerging medical treatments through continuing medical education. Delivering current and intelligible information to clinicians in a timely and non-time-consuming manner is the heart and challenge of continuing medical education.
About the Authors
Nisha Nair, MD
Dr. Nair is a talented medical writer with experience in the field of pharmacovigilance. Her experiences range from literary specialties such as single case processing, aggregate reporting, medical writing, and literature reviews, to industry focuses such as business development, team leadership, and client relationship management. Well versed in regulatory writing, Dr. Nair has conducted clinical trials training workshops for the biopharmaceutical industry and research institutions. Clients of Dr. Nair include established multinational companies as well as startups.
With proven people management skills, Dr. Nair has organized several successful projects and worked with people at all levels, motivating staff on both individual and team levels. For more information, contact her at nishanair@pacificsafetygroup.biz and view her LinkedIn page: https://www.linkedin.com/in/dr-nisha-n-puthiyedath/
DeeAnn Visk, PhD
DeeAnn Visk, PhD, is a San Diego based medical writer. Pharmacogenetics, high throughput screening, immunohistochemistry, confocal microscopy, and oncology are some of her content area specialties. Projects Dr. Visk has worked on include: articles for trade periodicals, editing of manuscripts before submission for peer-editing, writing manuscripts for submission, and informational articles from conferences. As the president of a local non-profit, the Association for Women in Science for two years, Dr. Visk gained experience in managing a 200-plus member organization. An active member of the Clinical Pharmacogenetic Implementation Consortium (CPIC), Dr. Visk was instrumental in the initiation of the Dissemination Working Group within CPIC.
When not researching, writing, or editing, Dr. Visk enjoys hiking and visiting National Parks. Residing northeast of downtown San Diego, Dr. Visk is married with two kids with two very spoiled hens in her backyard. For more information on her experience, please read this webpage and visit her LinkedIn profile. Dr. Visk can be reached at deeannlwv@gmail.com.