The 2015 meeting on Lipids—focusing on their impact in cancer, metabolic, and inflammatory diseases—took place on Tuesday and Wednesday, May 12 and 13 at the Scripps Seaside Forum at UCSD’s Scripps Institute of Oceanography (SIO). With a beautiful venue and superb facilities, what more can you ask for? How about some really interesting science.
Lipids are generally thought of as fats. But in a biological system, they are much more. They include chemokines and other signaling molecules involved in signal transduction to and from the cell membrane. Metabolically, lipids also play an important role. Innovation in technology allow the study all the lipids in an organism (yeast, bacteria, or animal), leading to a new field of study: lipidomics. Once again, UCSD is on the cutting edge, with an established program and website in the field.
Michael Snyder, the keynote speaker, has subjected himself to a battery of “omic” studies including his personal genome, exosome, microbiome, epigenome, proteome, metabolome, transcriptome, auto-antibody-ome, as well as cytokines. Data from these samples comprise the “Snyerdome”. All this was done in the interest of personalized medicine. These studies were done not only at one time point, but over a range of times, making it longitudinal.
Mike sees the data providing insights into how to managing healthcare in healthy individuals to predict risk, diagnose, monitor, and treat the patient, in this case, himself.
“He has also combined different state-of–the-art “omics” technologies to perform the first longitudinal detailed integrative personal omics profile (iPOP) of person and used this to assess disease risk and monitor disease states for personalized medicine” (from lab website).
in the future, Mike sees genomes being sequenced before birth and all this information being channeled through your smart phone. Patients will also bear more responsibility for maintaining their health with all the information they have; they will need to learn to maintain a balanced life.
The next speaker, David Wishart, discussed how to link lipidomics to laboratory medicine. He noted that in the rationalization of translating basic research to something of value in the clinic, researcher often cite the possibility of developing a new:
- surgical technique
- invent a new medical device
- drug
- drug target
- medically important gene
- biomarker
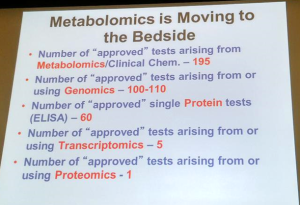
All these are good outcomes; some are more likely than others. Practitioners of lipidomics are most likely to have the best luck in developing new biomarkers; not many are surgeon and drug development has about a 0.001% success rate from basic science to the prescription bottle.
Discovery of new biomarkers is a realm where omics, specifically lipidomics, will meet a great chance of success. For this comparison, David recommends using the statistical ROC test, which is routinely used to evaluate medical test. This test gives a good sense of a medical test’s specificity and sensitivity by plotting the true positive rate over the false positive rate.
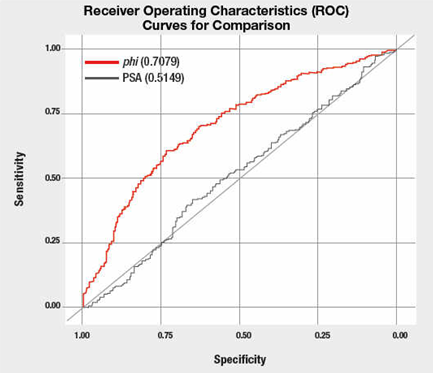
Example of ROC curve with an assessment of the area under the curve. The PSA referred to here is the amount of Prostate-Specific Antigen test; phi refers to a different, more specific calculation with less false positives than the PSA test alone.
Or you can just know that an ROC of 0.5 is worthless, while 1.0 is perfect.
Thus, from the graph above, using the PSA test alone to determine the risk of prostate cancer is poor. A better method is to use the phi method.
Work done looking at 3 to 5 biomarkers can have great ROC results. For example, predicting congenital heart defects by looking at the level of 3 carotenes, yields a ROC of 0.98. Other areas of success with high ROC scores include endometrial cancer, prostate cancer, and chronic fatigue syndrome.
David urged participants to become more quantitative to move their research into the clinic; using the website www.roccet.ca to generate ROC curves for your data is a great place to begin.
The numerous other speakers all gave fantastic talks.
In this smaller conference, I was able to browse through the all posters, read all the titles and talk to the presenters. Large conventions tend to lack the sense of intimacy and fraternity found in this lipidomics meeting. Kudos to the organizers for a successful event. A convivial group, I would highly recommend this meeting.